Author
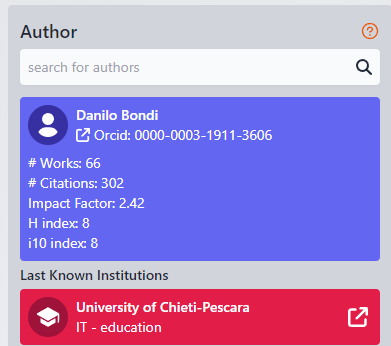 |
This part is giving the main information on the searched author.
Name Surname Orcid ID (click on to open Orcid page) # Works number # Citation NUmber Impact factor H Index i10 Index Last known Institute is the last published institute the author published; sometime you can have more because author decleared to works for all of them. |
|
An ORCID ID is a unique identifier for researchers, allowing them to distinguish their work from others with similar names. It ensures proper attribution of their publications, grants, and other academic contributions. The ORCID ID helps streamline the management of their scholarly record and enhances the visibility of their work.
| |
|
Impact factor
|
The Researcher Impact Factor measures the influence of a researcher's work by analyzing citation frequency and context. It indicates the significance and reach of their publications within the academic community, helping in assessments and funding decisions.
|
|
H Index
|
The H-Index measures a researcher's productivity and citation impact. It is defined as the number of papers (H) that have received at least H citations each. This metric helps evaluate the significance of a researcher’s cumulative contributions to their field.
|
|
i10 Index
|
The i10-Index indicates the number of a researcher's publications that have received at least 10 citations each. It is a simple metric used to gauge the impact and consistency of a researcher's work.
|
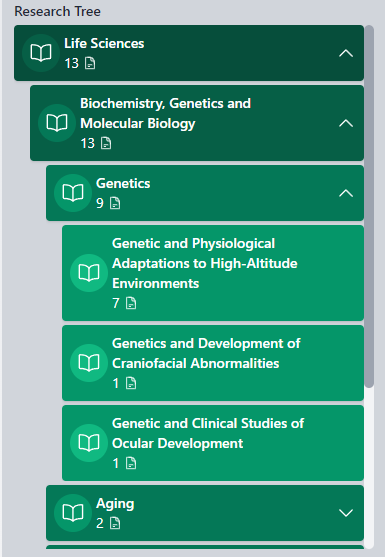 |
Research tree is the main areas researcher published by macroconcept(s). The Large Language Model here is extracting all data form author works, and after a computational classification assign these macro-categories.
Please note the number under the macroclass, it give an idea how is important (how many time LLM extracted and understood that clss) for the researcher this clss  With a mouse over, a filter icon let you filter only works with this class in
|
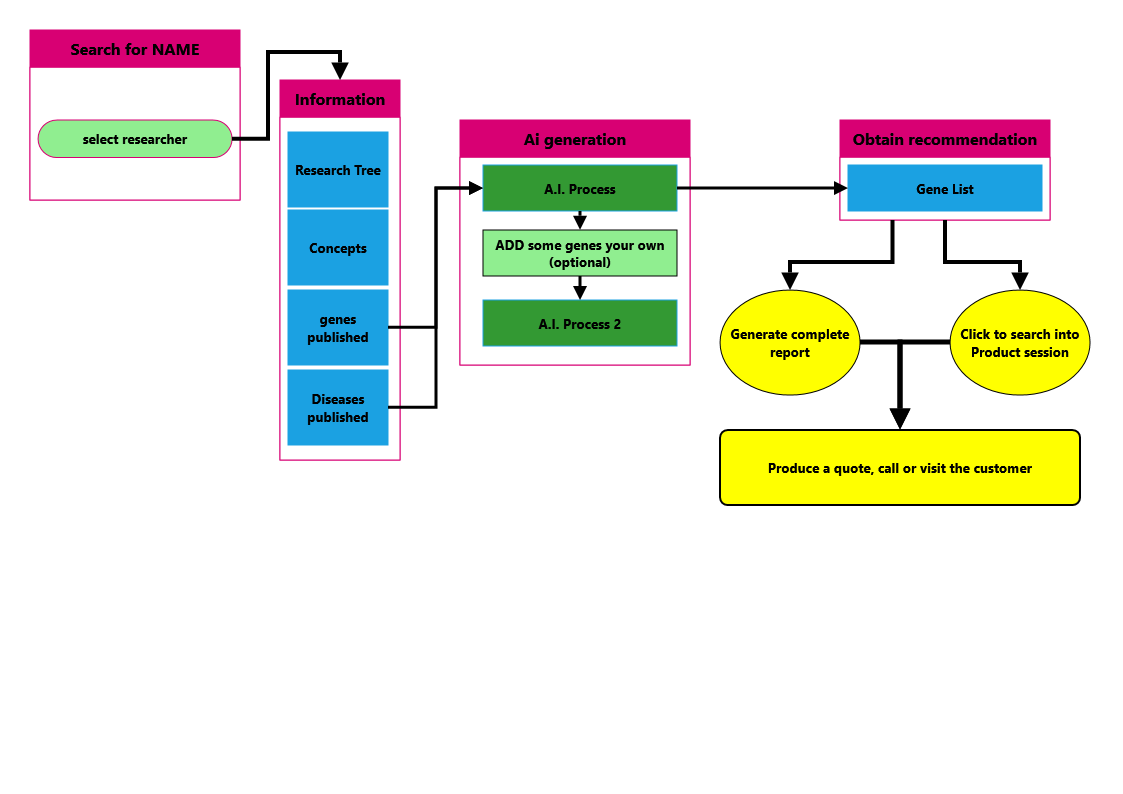
The author section is to search a specific name of a researcher, select and understand what he/she is doing to offer the proper products.
The typical schema for this section is
|
type a name, surname
|
select the right one
|
look to the publication list
apply a filter on date |
look to concepts, genes, disease published
|
look to genes
to recommend |
|
|
click
|
apply filter
|
click on icons to have more information
|
click on icons to have more information
|
 |
Click on Author section
|
|
Searching customers
| |
 | |
|
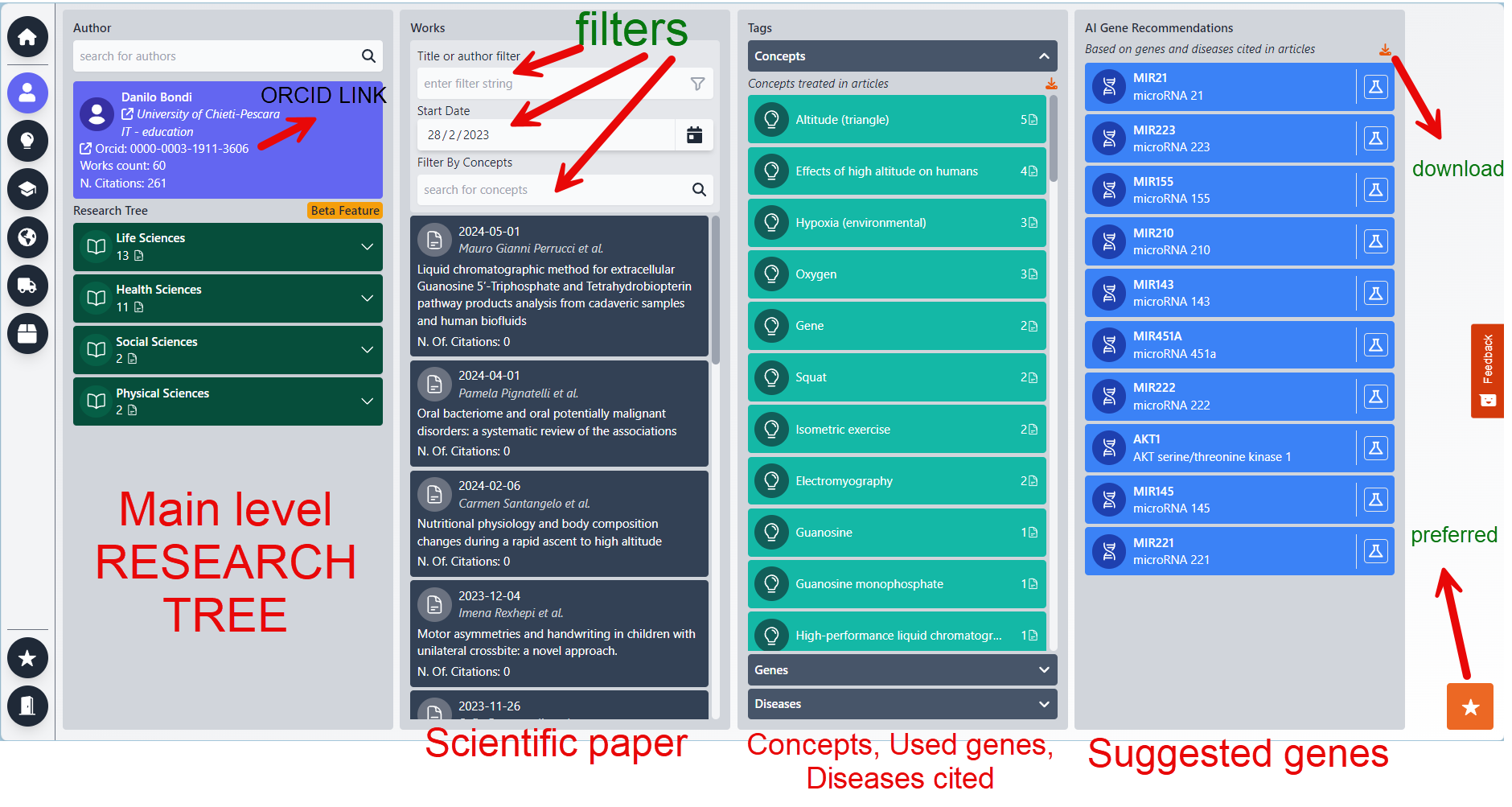
This section is designed for individual researchers, catering to scenarios such as preparing for a visit or gaining a general understanding of the researcher's work. It could also be used to facilitate an offer request. To begin, search for the researcher by name/surname and select the appropriate individual. Once selected, the BioRecommender system presents the following information:
| |
|
| |
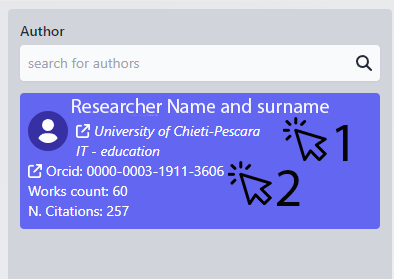 |
Column 1: The Author
Upon selecting a name from the list, additional options become available. You can click on the institution associated with the researcher's name and surname to open its details in a new browser tab. Alternatively, you can click on the researcher's ORCID ID to access more detailed information, including the possibility of obtaining their email address, provided there are sufficient permissions to do so.
ORCID: click on it to open the orcid website Works count: the number of scientific works for this author N.Citation: how many time these works have been cited |
 |
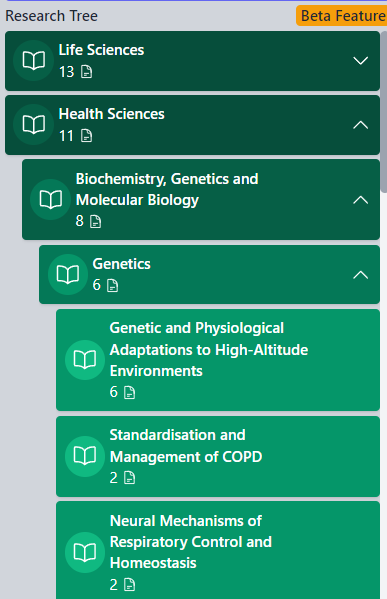
RESEARCH MAIN TREE
This section outlines the primary areas of research interest and the concepts that fall under them.
The hierarchy is structured into four levels, for example:
These categories are specifically related to the selected researcher and are provided to facilitate the exploration of other researchers who have contributed to similar topics in their work. This allows for further filtering by criteria such as country or institution.
 |
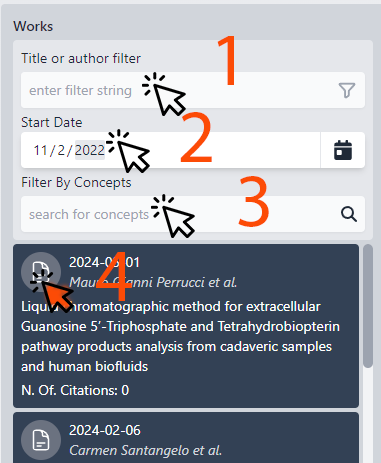 |
Column 2: The WORKS
This column displays all the published scientific works of the selected researcher.
1. Search and Filter: Locate works by title using the search function. You can then apply filters to narrow down the results.
2. Date Range: Adjust the date range to explore works published within specific timeframes. The list will update accordingly to reflect the selected period.
3. Concept Filter: Begin typing to generate a list of related concepts, allowing for more focused filtering of the works.
4. Accessing the Works: Click on a work to view its abstract or the full document, depending on the permissions granted by the researcher's licensing agreement.
Each entry provides a summary of the work, with the number of citations listed last. Works are sorted by their citation count, ensuring that the most influential publications are highlighted first.
|
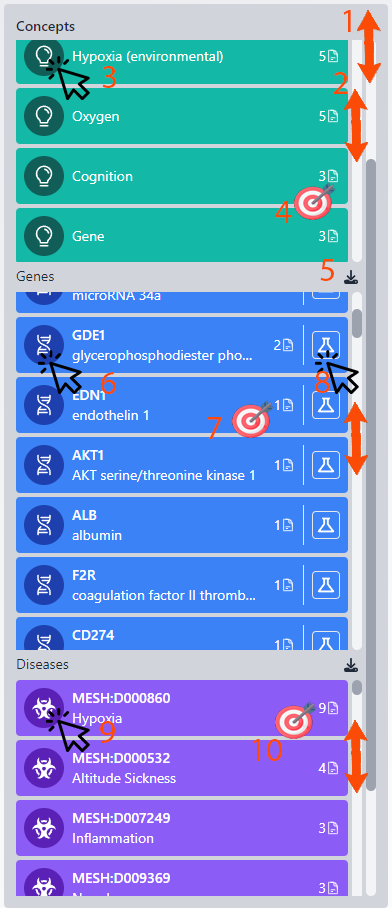 |
Column 3: Concepts/Genes/Diseases
This column is multifaceted, focusing on concepts, genes, and diseases related to the researcher's work. Please be aware of the two scrollbars: the first for navigating the entire column and the second within each subsection for scrolling through extensive lists.
1. Overall Column Scroll: Navigate through the entire column to view different sections.
2. Subsection Scroll: Each part of the column has its own scrollbar for when there are numerous records.
For Concepts: 3. Biorecommender Session: Initiating another session, the concept becomes the primary filter for finding related works, institutions, and authors. This feature enables searching for specific institutes (with country filters) related to the chosen concept, allowing for the exportation of a list of interested individuals.
4. Citation Count: Displays the number of times the researcher has cited a concept, with the count adjusting based on applied filters, such as selected time ranges.
For Genes: 5. Excel Export: Provides the option to download the section's data into an Excel file. A comprehensive author report feature is in development.
6. NCBI Link: Accesses the relevant gene page on www.ncbi.nlm.nih.gov for detailed protein information.
7. Gene Citation Count: Indicates the frequency of the author's citations of the gene within the filtered works.
For Diseases: 8. Product Matching: Connects genes to over 5 million products in the database, allowing selection by producer and direct linking to their websites. This section highlights genes cited by the author, suggesting potential future use based on their research focus and citation frequency.
9. Disease Information Link: Opens an external webpage with detailed information on the disease, including associated genes.
10. Disease Citation Count: Shows the number of times the disease has been cited by the author within the filtered timeframe, titles, or concepts.
|
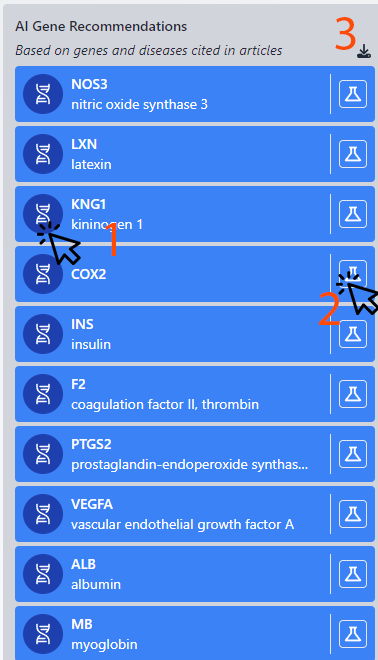 |
Column 4: Recommender Genes
This crucial section uses advanced neural network algorithms to predict genes that may be of interest to the author, based on their citation history in scientific works. The system analyzes the relationships between cited genes to identify potential new genes of relevance. Future enhancements will expand the capabilities of this feature.
For additional insights, our platform can also incorporate gene inquiries from customer requests. By leveraging an API, we can provide predictions on the most likely SKUs needed by customers.
1. NCBI Gene Information: Access detailed gene data through an external link to the NCBI database, offering a comprehensive overview.
2. Product Matching: This function aligns selected genes with over 5 million products in our database, facilitating the selection of products by producer and providing direct links to their websites.
3. Excel Download: Users can download a curated list of recommended genes in Excel format for further analysis or record-keeping.
It's important to note that this column highlights genes previously cited (and possibly utilized) by the author. This implies a likelihood of recurring use in similar areas of study, especially if a gene is frequently cited, indicating its significance in the researcher's work.
|
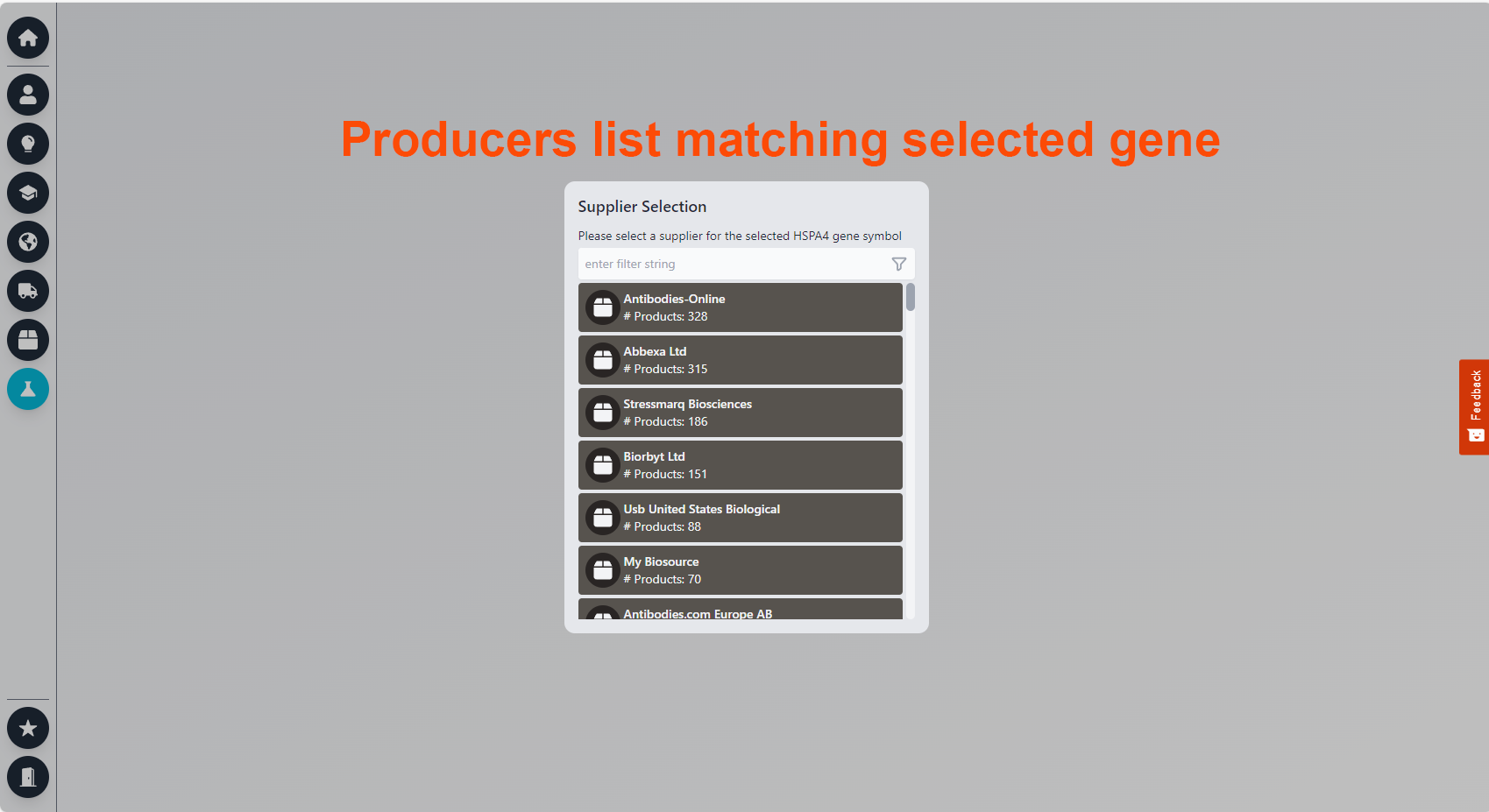 |
You clicked on the flask, and you are now in the product section
|
 | |
|
| |
 | |
